Background
For years I’ve been contributing to and maintaining, at work, a software stack for streamflow forecasting. It is a stack with a core in C++, but accessible via a C API by users from R, Matlab, Python and so on. A whole article could be written about the design rationale, successes and shortcomings of this stack, and the interplay of people, organisations and technologies in using these tools and how. But this will not be this post.
Focussing on the Python side of things, these streamflow forecasting tools are used mostly on Windows and Linux, the core is deployed as dynamic libraries (.dll or .so) on disk, and python packages access these using cffi for interoperability. The python packages contain solely python code; there is no cython or straight C.
I’ve come to appreciate (mostly) conda environments for managing software stacks for various projects. This post is a start to test packaging some of “my” software with conda, in the hope this reduces the surprisingly strong impedance, technical but not only, towards usage by a broader audience.
A bit picture end point would be a corporate equivalent to a conda-forge channel, with the full software stack available for any employee.
Baby steps
I had a look a few weeks ago at how I’d package a substantial but relatively small C++ code MOIRAI: Manage C++ Objects’s lifetime when exposed through a C API. This proved a bit premature for reasons I won’t detail.
So, let’s (re)start with a python-only library, as it happens in the same vein, refcount. Astonishingly, there is still no strict equivalent that I can find in conda-forge dedicated to reference counting external pointers. So, can I claim the spot?
Resources
I started this post thinking first about conda packaging rather than submission per se to conda-forge. Some resources I initially looked at as promising, but from which I backed away (for now):
- Building conda packages from scratch
- python packaging tutorials Scipy 2018 Tutorial: The Joy of Packaging - conda packages
- Activision Game: A tutorial (+ build recipes) describing how to use conda for C, C++, and python package management outlines well the rationale for packaging in conda, and appears didactic. It appears not to have recent commit, although it is not necessarily a problem.
You’ll see in the walkthrough below (next section) that I reoriented towards an upfront submission to conda-forge:
- 3 simple stept to build a python package for conda-forge. This post really got me on a better path.
- conda-forge documentation: Contributing packages
Walkthrough
First trial: conda build locally?
Of course python is necessary and a conda environment a given. I do . ~/config/baseconda because I never have conda activated by default from .bashrc.
I am actually not sure from the conda-build tutorial in which environment I should install conda-build. Let’s try the base environment and see whether we get stuck or not.
mamba install -c conda-forge conda-build (installing from conda-forge is an idiosyncrasy. I strongly recommend mamba)
You should at least skim through the concepts. This is rather dry to read throughly upfront.
The tutorial Building conda packages from scratch quickly confused me; I was trying to transpose it to refcount but this does not look like the right template to start from. The section editing-the-meta-yaml-file appears out of sync with the “correct” meta.yaml file. Baffling.
Preparing a PR to conda-forge/staged-recipes
Enter two new resources: 3 simple stept to build a python package for conda-forge and conda-forge documentation: Contributing packages. From these it becomes clear I should use grayskull to get a starting point as a meta.yaml file
cd ~/src
git clone --depth 1 git@github.com:jmp75/staged-recipes.git
cd ~/src/staged-recipes/recipes
git remote add upstream git@github.com:conda-forge/staged-recipes.git
mamba install -c conda-forge grayskull
grayskull pypi refcount#### Initializing recipe for refcount (pypi) ####
Recovering metadata from pypi...
Starting the download of the sdist package refcount
refcount 100% Time: 0:00:00 15.3 MiB/s|###############################################################################################################################################################################################################################################|
Recovering information from setup.py
Executing injected distutils...
Recovering metadata from setup.cfg
No data was recovered from setup.py. Forcing to execute the setup.py as script
Recovering metadata from setup.cfg
Checking >> cffi 100% |##########################################################################################################################################################################################################################################|[Elapsed Time: 0:00:00]
Matching license file with database from Grayskull...
Match percentage of the license is 59%. Low match percentage could mean that the license was modified.
License type: BSD-3-Clause
License file: LICENSE.txt
Host requirements:
- pip
- python
Run requirements:
- cffi
- python
RED: Missing packages
GREEN: Packages available on conda-forge
Maintainers:
- j-m
#### Recipe generated on /home/abcdef/src/staged-recipes/recipes for refcount ####The output meta.yaml (which is actually a ninja template file), is a good start, however you should revise it a bit rather than accept wholesale
Mostly fine, however this did not pick up a requirement cffi >=1.11.5, and second guessing from reading this gallon.me post, a minimum python version is necessary to get accepted.
requirements:
host:
- pip
- python >=3.6
run:
- cffi >=1.11.5
- python >=3.6Perhaps optional, remove a hard-coded package string “refcount”in the source url section.
It is instructive to look at the existing pull requests on staged-recipes. Notably I realise that the github ID extracted by grayskull is not the correct one; I am not the ID j-m, unfortunately (could have been judging by history length).
extra:
recipe-maintainers:
- j-mextra:
recipe-maintainers:
- jmp75The end result should be something like:
{% set name = "refcount" %}
{% set version = "0.9.3" %}
package:
name: {{ name|lower }}
version: {{ version }}
source:
url: https://pypi.io/packages/source/{{ name[0] }}/{{ name }}/{{ name }}-{{ version }}.zip
sha256: bf8bfabdac6f0d9fe3734f1c1830fda8b9b2d740c90ecf8caf8c2ef3ed9c8442
build:
noarch: python
script: {{ PYTHON }} -m pip install . -vv
number: 0
requirements:
host:
- pip
- python >=3.6
run:
- cffi >=1.11.5
- python >=3.6
test:
imports:
- refcount
commands:
- pip check
requires:
- pip
about:
home: https://github.com/csiro-hydroinformatics/pyrefcount
summary: A Python package for reference counting and interop with native pointers
description: |
This package helps you achieve reliable management of memory
allocated in native libraries, written for instance in C++. While
it boils down to "simply" maintaining a set of counters,
it is deceptively complicated to do so properly and not end up
with memory leaks or crashes.
<https://pyrefcount.readthedocs.io/en/latest/>.
dev_url: https://github.com/csiro-hydroinformatics/pyrefcount
license: BSD-3-Clause
license_family: BSD
license_file: LICENSE.txt
extra:
recipe-maintainers:
- jmp75So; ready to submit a pull request? Wait, wait.
Building locally
test has a section on Running unit tests. Note that the default conda recipe above has a “pip check”, but nothing more. refcount unit tests use pytest, and has unit tests; tick that. refcount is a pure python package, but it is a package for (mostly) interoperability with native code via a C API. Unit tests do contain some c/c++ code.
Should the recipe run fine upon submission, including unit tests? Before submitting a pull request that may trigger a failed check, let’s experiment with staging tests locally.
so:
cd ~/src/staged-recipes
python ./build-locally.py linux64 File "/home/abcdef/miniconda/lib/python3.9/subprocess.py", line 373, in check_call
raise CalledProcessError(retcode, cmd)
subprocess.CalledProcessError: Command '['.scripts/run_docker_build.sh']' returned non-zero exit status 1.I tried to conda install -c conda-forge shyaml which seems to be used by the scripts, but this did not alleviate the issue.
That took me some time to find a workaround to this one. The “exit status 1” is actually very misleading. The root cause is a docker run -it that exited with an error code 139. I seem to not be the only one to have bumped into this issue still open. I may have pointed to the workaround in the conda-forge FAQ.
I needed to override the default docker image build-locally.py falls back to with:
export DOCKER_IMAGE=quay.io/condaforge/linux-anvil-cos7-x86_64
python build-locally.py linux64the build script works this time, but at some point:
Processing $SRC_DIR
Added file://$SRC_DIR to build tracker '/tmp/pip-build-tracker-bkn7ckr9'
Running setup.py (path:$SRC_DIR/setup.py) egg_info for package from file://$SRC_DIR
Created temporary directory: /tmp/pip-pip-egg-info-68li1y6t
Preparing metadata (setup.py): started
Running command python setup.py egg_info
Traceback (most recent call last):
File "<string>", line 2, in <module>
File "<pip-setuptools-caller>", line 34, in <module>
File "/home/conda/staged-recipes/build_artifacts/refcount_1654312313810/work/setup.py", line 41, in <module>
with open(os.path.join(here, 'README.md'), encoding='utf-8') as f:
File "/home/conda/staged-recipes/build_artifacts/refcount_1654312313810/_h_env_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_placehold_pl/lib/python3.10/codecs.py", line 905, in open
file = builtins.open(filename, mode, buffering)
FileNotFoundError: [Errno 2] No such file or directory: '/home/conda/staged-recipes/build_artifacts/refcount_1654312313810/work/README.md'
error: subprocess-exited-with-error
× python setup.py egg_info did not run successfully.
│ exit code: 1
╰─> See above for output.
note: This error originates from a subprocess, and is likely not a problem with pip.
This is an issue that may be in my control.
cd ~/src/staged-recipes/build_artifacts/refcount_1654312313810/work
ls
## build_env_setup.sh conda_build.sh LICENSE.txt MANIFEST.in metadata_conda_debug.yaml PKG-INFO README.rst refcount refcount.egg-info setup.cfg setup.pyrefcount has both a README.md and README.rst, the latter being an export from the former because pypi requires (or used to require) a README.rst to display correctly. The zip archive of the source code on pypi indeed does not have the README.md file included.
I’ve inherited the practice to use in the packages setup.py the following, to limit redundances.
with open(os.path.join(here, 'README.md'), encoding='utf-8') as f:
long_description = f.read()
long_description_content_type='text/markdown'Previously I needed to convert on the fly to restructured Text, but Markdown is more supported. Still there is a lot of inertia with restructuredText.
I may try to just nuke the README.rst. The only fly on the ointment is: is pypi ok with rendering markdown correctly these days? Probably; the packaging documentation is using README.md by default.
So, build and submit to pypi the updated refcount 0.9.4 with no README.rst. Looks fine, including the zip source archive.
RuntimeError: SHA256 mismatch: '21567918cb1bb30bf8116ce3483d3f431de202618eabbc6887b4814b40a3b94a' != 'bf8bfabdac6f0d9fe3734f1c1830fda8b9b2d740c90ecf8caf8c2ef3ed9c8442'
Traceback (most recent call last):Right, I forgot to change the checksum in the meta.yaml file.
And… it seems to complete.
import: 'refcount'
+ pip check
No broken requirements found.
+ exit 0
Resource usage statistics from testing refcount:
Process count: 1
CPU time: Sys=0:00:00.0, User=-
Memory: 3.0M
Disk usage: 28B
Time elapsed: 0:00:02.1
TEST END: /home/conda/staged-recipes/build_artifacts/noarch/refcount-0.9.4-pyhd8ed1ab_0.tar.bz2Submit the pull request, and pleasantly:
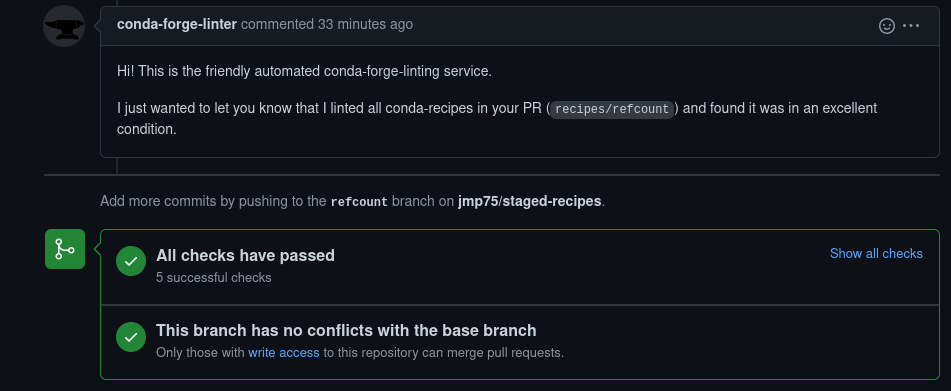
Conclusion
While there were a couple of bumps along the way, this should end up with a positive outcome. If not with refcount on conda-forge, I’ve a better understanding to tackle conda packaging on the rest of the software stack.
- Building a conda package “from scratch” may not be the easiest learning path. Even if you indent to build a conda package not for conda-forge, going through the staged-recipes process may be a most
- Some of the reference documentation may need a spruice up. Building conda packages from scratch confused me. First, packaging a pypi package is not starting “from scratch” for most users. Second, inconsistencies in the documentation. I am sure I’ll get back to that resource, but I wish there were more “water tight”, step-by-step tutorials for conda packaging.