import pandas as pd
import numpy as np
import xarray as xr
from swift2.simulation import create_subarea_simulation, create_catchment
# etc. etc, other imports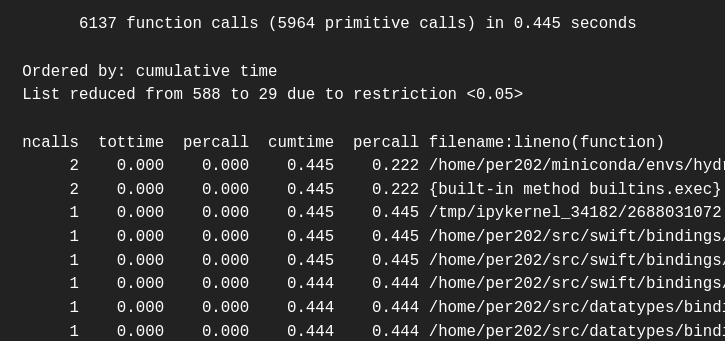
Background
A software stack for streamflow forecasting consists of a C++ modelling engine, accessible via a C API from various interactive language bindings including python. I have not done some benchmark performance profiling for a while on this stack. I expect the C++ core to be pretty much as good as it was 8 years ago, but as I bed down some of the python bindings, I notice some lags when interacting. This is not a surprise, and I expect these to be either in python code, or perhaps at the boundary with the C API. This intuition needs to be confirmed by objective measurements.
It is one of these things that are rarely a blocker for quite a while, but a bit of instrumentation and performance tuning improves the user experience, a little bit every day. Also, some use cases where substantial stochastic data is generated in Python and passed to the C++ core would be very penalised by inefficiencies in Python or Python/C interoperability code.
Foreword: in the case below, we do not need to profile C++ code per se after all, so if this is what you are after specifically. Read on for the Python side of the story.
Python profilers
9 fine libraries for profiling python code is a recent article, as I write. Of these, Palanteer is interesting in its mixed mode profiling capabilities (Python and C++). I’ll have to explore it, perhaps not just yet though. A priori the cProfiler coming with the Python base library is all I need for this immediate use case.
Runtime profiling
I will skip on the details of the imported libraries here.
Creating synthethic hydrologic model
The overall purpose of the exercise will be to measure performance under various conditions: model structure, size, time steps, etc. We will not do all that in this post; suffice to say we define a set of functions to create synthetic model setups. We do not show the full definitions. To give a flavour
from cinterop.timeseries import mk_hourly_xarray_series
def create_pulses(nTimeSteps, start) : return mk_hourly_xarray_series( createPulse(nTimeSteps, 5.0, 48), start)
def create_uniform(nTimeSteps, start) : return mk_hourly_xarray_series( createUniform(nTimeSteps, 1.0), start)
def set_test_input(ms:'Simulation', subAreaName:'str', rainVarName, petVarName, rain_ts, pet_ts):
p_id = mk_full_data_id('subarea', subAreaName, rainVarName)
e_id = mk_full_data_id('subarea', subAreaName, petVarName)
ms.play_input( rain_ts, p_id)
ms.play_input( pet_ts, e_id)
# def create_line_system(n_time_steps, n_links, rain_varname = "P", pet_varname = "E", area_km2 = 1):
# et caetera We are now ready to create our catchment simulation. Before we plunge into cProfiler let’s use a simpler way to assess the runtime from notebooks:
Using notebook’s %%time
Let’s create a setup with 15 subareas, hourly time step over 10 years.
%%time
n_time_step = 10 * 365 * 24
ms = create_line_system(n_time_step, 15)CPU times: user 888 ms, sys: 8.94 ms, total: 897 ms
Wall time: 897 msWell, this was only the create of the baseline model, not even execution, and this already takes close to a second. Granted, there are a fair few hours in a decade. Still, a whole second!
What about the simulation runtime? Let’s parameterise minimally to avoid possible artefacts, and execute.
from swift2.doc_helper import configure_hourly_gr4j, get_free_params
configure_hourly_gr4j(ms)
p = create_parameteriser('Generic subarea', get_free_params("GR4J"))
p.apply_sys_config(ms)Double check we are indeed running hourly over 10 years:
ms.get_simulation_span(){'start': datetime.datetime(1989, 1, 1, 0, 0),
'end': datetime.datetime(1998, 12, 29, 23, 0),
'time step': 'hourly'}%%time
ms.exec_simulation()CPU times: user 314 ms, sys: 691 µs, total: 315 ms
Wall time: 314 msThis is actually quite good, and “unexpectedly” less than the model creation itself. This is actually not all that surprising. All of the model execution happens in C++ land. The model setup involves much more operations in python.
Let’s look at an operation exchanging data from the C++ engine for display in Python. The model simulation has some of its states receiving input time series:
ms.get_played_varnames()[:6]['subarea.1.E',
'subarea.1.P',
'subarea.10.E',
'subarea.10.P',
'subarea.11.E',
'subarea.11.P']Let’s see what happens in the retrieval of one of these input time series:
%%time
ts = ms.get_played("subarea.1.E")CPU times: user 441 ms, sys: 106 µs, total: 441 ms
Wall time: 440 msThis is substantial; more than the native execution over a catchment with 15 subareas. So:
- Can we identify the hotspot(s)?
- Can we do something to improve it.
Profiling
Enter cProfile, as we will stick with this in this post. Adapting some of the sample code shown in the Python documentation on profilers
import cProfileimport pstats, io
pr = cProfile.Profile()
pr.enable()
ts = ms.get_played("subarea.1.E")
pr.disable()s = io.StringIO()
sortby = pstats.SortKey.CUMULATIVE
ps = pstats.Stats(pr, stream=s).sort_stats(sortby)We will print only the top 5 % of the list of function calls, and see if we can spot the likely hotspot
ps.print_stats(.05)
print(s.getvalue()) 6137 function calls (5964 primitive calls) in 0.445 seconds
Ordered by: cumulative time
List reduced from 588 to 29 due to restriction <0.05>
ncalls tottime percall cumtime percall filename:lineno(function)
2 0.000 0.000 0.445 0.222 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/IPython/core/interactiveshell.py:3361(run_code)
2 0.000 0.000 0.445 0.222 {built-in method builtins.exec}
1 0.000 0.000 0.445 0.445 /tmp/ipykernel_34182/2688031072.py:4(<cell line: 4>)
1 0.000 0.000 0.445 0.445 /home/abcdef/src/swift/bindings/python/swift2/swift2/classes.py:471(get_played)
1 0.000 0.000 0.445 0.445 /home/abcdef/src/swift/bindings/python/swift2/swift2/play_record.py:190(get_played)
1 0.000 0.000 0.444 0.444 /home/abcdef/src/swift/bindings/python/swift2/swift2/internal.py:94(internal_get_played_tts)
1 0.000 0.000 0.444 0.444 /home/abcdef/src/datatypes/bindings/python/uchronia/uchronia/data_set.py:10(get_multiple_time_series_from_provider)
1 0.000 0.000 0.444 0.444 /home/abcdef/src/datatypes/bindings/python/uchronia/uchronia/internals.py:69(internal_get_multiple_time_series)
1 0.000 0.000 0.441 0.441 /home/abcdef/src/datatypes/bindings/python/uchronia/uchronia/internals.py:76(<listcomp>)
1 0.000 0.000 0.441 0.441 /home/abcdef/src/datatypes/bindings/python/uchronia/uchronia/internals.py:74(f)
1 0.000 0.000 0.441 0.441 /home/abcdef/src/datatypes/bindings/python/uchronia/uchronia/internals.py:79(internal_get_single_model_time_series)
1 0.002 0.002 0.441 0.441 /home/abcdef/src/swift/bindings/python/swift2/swift2/wrap/swift_wrap_custom.py:216(get_played_pkg)
1 0.000 0.000 0.438 0.438 /home/abcdef/src/c-interop/bindings/python/cinterop/cinterop/cffi/marshal.py:386(geom_to_xarray_time_series)
1 0.000 0.000 0.368 0.368 /home/abcdef/src/c-interop/bindings/python/cinterop/cinterop/cffi/marshal.py:367(ts_geom_to_even_time_index)
1 0.000 0.000 0.368 0.368 /home/abcdef/src/c-interop/bindings/python/cinterop/cinterop/timeseries.py:22(create_even_time_index)
1 0.368 0.368 0.368 0.368 /home/abcdef/src/c-interop/bindings/python/cinterop/cinterop/timeseries.py:25(<listcomp>)
16 0.000 0.000 0.071 0.004 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/xarray/core/dataarray.py:365(__init__)
1 0.000 0.000 0.070 0.070 /home/abcdef/src/c-interop/bindings/python/cinterop/cinterop/cffi/marshal.py:356(create_ensemble_series)
9 0.000 0.000 0.070 0.008 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/xarray/core/variable.py:74(as_variable)
1 0.000 0.000 0.070 0.070 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/xarray/core/dataarray.py:90(_infer_coords_and_dims)
36 0.000 0.000 0.070 0.002 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/xarray/core/variable.py:181(as_compatible_data)
35/31 0.060 0.002 0.060 0.002 {built-in method numpy.asarray}
1 0.000 0.000 0.009 0.009 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/xarray/core/variable.py:172(_possibly_convert_objects)
1 0.000 0.000 0.009 0.009 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/pandas/core/series.py:323(__init__)
2 0.000 0.000 0.009 0.005 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/pandas/core/construction.py:470(sanitize_array)
2 0.000 0.000 0.009 0.005 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/pandas/core/construction.py:695(_try_cast)
1 0.000 0.000 0.009 0.009 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/pandas/core/dtypes/cast.py:1466(maybe_infer_to_datetimelike)
2 0.000 0.000 0.006 0.003 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/pandas/core/arrays/datetimes.py:1994(_sequence_to_dt64ns)
1 0.000 0.000 0.006 0.006 /home/abcdef/miniconda/envs/hydrofc/lib/python3.9/site-packages/pandas/core/dtypes/cast.py:1499(try_datetime)
in a file c-interop/bindings/python/cinterop/cinterop/timeseries.py the function create_even_time_index appears to be where a lengthy operation occurs. More precisely, when this function does a list comprehension (listcomp). Note that I infer this because in the cumtime (cumulative time) column there is a drop from 0.396 ms for listcomp to 0.071 for the rest of the operations under this function. I think this is the right way to interpret it in this case, but it may not be the case in other profiling context.
The code for create_even_time_index is at this permalink
def create_even_time_index(start:ConvertibleToTimestamp, time_step_seconds:int, n:int) -> List:
start = as_timestamp(start)
delta_t = np.timedelta64(time_step_seconds, 's')
return [start + delta_t * i for i in range(n)][start + delta_t * i for i in range(n)] is the bulk of the time, .396 out of a total 0.477 ms.
This list is created as the basis for a time index for the creation of the xarray object returned by the overall get_played function. So, is there a faster way to create this time series index?
Performance tuning
start = pd.Timestamp(year=2000, month=1, day=1)n= 24*365*10def test_index_creation(start, n:int) -> List:
start = as_timestamp(start)
time_step_seconds = 3600
delta_t = np.timedelta64(time_step_seconds, 's')
return [start + delta_t * i for i in range(n)]%%time
a = test_index_creation(start, n)CPU times: user 352 ms, sys: 562 µs, total: 353 ms
Wall time: 351 msstart is a pandas Timestamp, and we add to it an object of type np.timedelta64 87600 times. I doubt this is the main issue, but let’s operate in numpy types as much as we can by converting the pd.Timestamp once:
startTimestamp('2000-01-01 00:00:00')start.to_datetime64()numpy.datetime64('2000-01-01T00:00:00.000000000')def test_index_creation(start, n:int) -> List:
start = as_timestamp(start).to_datetime64()
time_step_seconds = 3600
delta_t = np.timedelta64(time_step_seconds, 's')
return [start + delta_t * i for i in range(n)]%%time
a = test_index_creation(start, n)CPU times: user 293 ms, sys: 8.48 ms, total: 301 ms
Wall time: 300 msThis is actually more of an improvement than I anticipated. OK. What else can we do?
Pandas has the helpful Time series / date functionality page in its documentation. The function from which we started is generic, but for important cases such as hourly and daily time steps, there are options to use the freq argument to the date_range function
%%time
pd.date_range(start, periods=n, freq="H")CPU times: user 242 µs, sys: 686 µs, total: 928 µs
Wall time: 520 µsDatetimeIndex(['2000-01-01 00:00:00', '2000-01-01 01:00:00',
'2000-01-01 02:00:00', '2000-01-01 03:00:00',
'2000-01-01 04:00:00', '2000-01-01 05:00:00',
'2000-01-01 06:00:00', '2000-01-01 07:00:00',
'2000-01-01 08:00:00', '2000-01-01 09:00:00',
...
'2009-12-28 14:00:00', '2009-12-28 15:00:00',
'2009-12-28 16:00:00', '2009-12-28 17:00:00',
'2009-12-28 18:00:00', '2009-12-28 19:00:00',
'2009-12-28 20:00:00', '2009-12-28 21:00:00',
'2009-12-28 22:00:00', '2009-12-28 23:00:00'],
dtype='datetime64[ns]', length=87600, freq='H')It is two order of magnitude faster… Definitely worth a re-engineering of the features in the timeseries.py we started from.
This probably does not solve the issue for many other cases (irregular time steps, e.g. monthly), but there are many cases where we could benefit. The date_range documentation specifies that an arbitrary DateOffset to its freq argument (freq: str or DateOffset, default ‘D’). How efficient is this operation on our 87600 data points?
d_offset = pd.tseries.offsets.DateOffset(minutes=15)start + d_offsetTimestamp('2000-01-01 00:15:00')%%time
pd.date_range(start, periods=n, freq=d_offset)CPU times: user 1.5 s, sys: 1.32 ms, total: 1.5 s
Wall time: 1.5 sDatetimeIndex(['2000-01-01 00:00:00', '2000-01-01 00:15:00',
'2000-01-01 00:30:00', '2000-01-01 00:45:00',
'2000-01-01 01:00:00', '2000-01-01 01:15:00',
'2000-01-01 01:30:00', '2000-01-01 01:45:00',
'2000-01-01 02:00:00', '2000-01-01 02:15:00',
...
'2002-07-01 09:30:00', '2002-07-01 09:45:00',
'2002-07-01 10:00:00', '2002-07-01 10:15:00',
'2002-07-01 10:30:00', '2002-07-01 10:45:00',
'2002-07-01 11:00:00', '2002-07-01 11:15:00',
'2002-07-01 11:30:00', '2002-07-01 11:45:00'],
dtype='datetime64[ns]', length=87600, freq='<DateOffset: minutes=15>')It looks like in this case this is actually a fair bit slower than my original implementation. Interesting. And using start.to_datetime64() makes no difference too.
Conclusion
This demonstrated a relatively simple, but real case where cProfiler helps alleviate usually small but pervasive runtime inefficiencies. In this case, so far we have not needed to look close to the Python/C interoperability layer. The key bottleneck was pure python. I envisage I may post something later on looking at trickier situation.
To be honest, I did second guess a few things upfront. But the cProfiler and simpler %%time brought at least confirmation and sometimes useful, conter-intuitive insight. “You cannot manage what you cannot measure” as the saying goes.